Intermediate Python
Modularization and Documentation
Modularization and Documentation
Now that we've covered some of the basic syntax and libraries in Python we can start to tackle our data analysis problem. We are interested in understanding the relationship between the weather and the number of mosquitos so that we can plan mosquito control measures. Since we want to apply these mosquito control measures at a number of different sites we need to understand how the relationship varies across sites. Remember that we have a series of CSV files with each file containing the data for a single location.
Learning Objectives
- Write code for people, not computers
- Break a program into chunks
- Write and use functions in Python
- Write useful documentation
Starting small
When approaching computational tasks like this one it is typically best to start small, check each piece of code as you go, and make incremental changes. This helps avoid marathon debugging sessions because it's much easier to debug one small piece of the code at a time than to write 100 lines of code and then try to figure out all of the different bugs in it.
Let's start by reading in the data from a single file and conducting a simple regression analysis on it. In fact, I would actually start by just importing the data and making sure that everything is coming in OK.
import pandas as pd
d = pd.read_csv('A2_mosquito_data.csv')
print d year temperature rainfall mosquitos
0 1960 82 200 180
1 1961 70 227 194
2 1962 89 231 207
3 1963 74 114 121
4 1964 78 147 140
5 1965 85 151 148
6 1966 86 172 162
7 1967 75 106 112
8 1968 70 276 230
9 1969 86 165 162
10 1970 83 222 198
11 1971 78 297 247
12 1972 87 288 248
13 1973 76 286 239
14 1974 86 231 202
15 1975 90 284 243
16 1976 76 190 175
17 1977 87 257 225
18 1978 88 128 133
19 1979 87 218 199
20 1980 81 206 184
21 1981 74 175 160
22 1982 85 202 187
23 1983 71 130 126
24 1984 80 225 200
25 1985 72 196 173
26 1986 76 261 222
27 1987 85 111 121
28 1988 83 247 210
29 1989 86 137 142
30 1990 82 159 152
31 1991 77 172 160
32 1992 74 280 231
33 1993 70 291 238
34 1994 77 126 125
35 1995 89 191 178
36 1996 83 298 248
37 1997 80 282 237
38 1998 86 219 195
39 1999 72 143 134
40 2000 79 262 221
41 2001 85 189 175
42 2002 86 205 186
43 2003 72 195 173
44 2004 78 148 146
45 2005 71 262 219
46 2006 88 255 226
47 2007 79 262 221
48 2008 73 198 176
49 2009 86 215 187
50 2010 87 127 129
The import seems to be working properly, so that's good news, but does anyone have anyone see anything about the code that they don't like?
That's right. The variable name I've chosen for the data doesn't really communicate any information to anyone about what it's holding, which means that when I come back to my code next month to change something I'm going to have a more difficult time understanding what the code is actually doing. This brings us to one of our first major lessons for the morning, which is that in order to understand what our code is doing so that we can quickly make changes in the future, we need to write code for people, not computers, and an important first step is to use meaningful varible names.
import pandas as pd
data = pd.read_csv('A2_mosquito_data.csv')
print data.head() year temperature rainfall mosquitos
0 1960 82 200 180
1 1961 70 227 194
2 1962 89 231 207
3 1963 74 114 121
4 1964 78 147 140
The .head() method lets us just look at the first few rows of the data. A method is a function attached to an object that operates on that object. So in this case we can think of it as being equivalent to head(data).
Everything looks good, but either global warming has gotten really out of control or the temperatures are in degrees Fahrenheit. Let's convert them to Celcius before we get started.
We don't need to reimport the data in our new cell because all of the executed cells in IPython Notebook share the same workspace. However, it's worth noting that if we close the notebook and then open it again it is necessary to rerun all of the individual blocks of code that a code block relies on before continuing. To rerun all of the cells in a notebook you can select Cell -> Run All from the menu.
data['temperature'] = (data['temperature'] - 32) * 5 / 9.0
print data.head() year temperature rainfall mosquitos
0 1960 27.777778 200 180
1 1961 21.111111 227 194
2 1962 31.666667 231 207
3 1963 23.333333 114 121
4 1964 25.555556 147 140
That's better. Now let's go ahead and conduct a regression on the data. We'll use the statsmodels library to conduct the regression.
import statsmodels.api as sm
regr_results = sm.OLS.from_formula('mosquitos ~ temperature + rainfall', data).fit()
regr_results.summary()<class 'statsmodels.iolib.summary.Summary'>
"""
OLS Regression Results
==============================================================================
Dep. Variable: mosquitos R-squared: 0.997
Model: OLS Adj. R-squared: 0.997
Method: Least Squares F-statistic: 7889.
Date: Sat, 18 Jan 2014 Prob (F-statistic): 3.68e-61
Time: 13:10:18 Log-Likelihood: -111.54
No. Observations: 51 AIC: 229.1
Df Residuals: 48 BIC: 234.9
Df Model: 2
===============================================================================
coef std err t P>|t| [95.0% Conf. Int.]
-------------------------------------------------------------------------------
Intercept 17.5457 2.767 6.341 0.000 11.983 23.109
temperature 0.8719 0.092 9.457 0.000 0.687 1.057
rainfall 0.6967 0.006 125.385 0.000 0.686 0.708
==============================================================================
Omnibus: 1.651 Durbin-Watson: 1.872
Prob(Omnibus): 0.438 Jarque-Bera (JB): 0.906
Skew: -0.278 Prob(JB): 0.636
Kurtosis: 3.343 Cond. No. 1.92e+03
==============================================================================
Warnings:
[1] The condition number is large, 1.92e+03. This might indicate that there are
strong multicollinearity or other numerical problems.
"""As you can see statsmodels lets us use the names of the columns in our dataframe to clearly specify the form of the statistical model we want to fit. This also makes the code more readable since the model we are fitting is written in a nice, human readable, manner. The summary method gives us a visual representation of the results. This summary is nice to look at, but it isn't really useful for doing more computation, so we can look up particular values related to the regression using the regr_results attributes. These are variables that are attached to regr_results.
print regr_results.params
print regr_results.rsquaredIntercept 17.545739
temperature 0.871943
rainfall 0.696717
dtype: float64
0.996966873691
If we want to hold onto these values for later we can assign them to variables:
parameters = regr_results.params
rsquared = regr_results.rsquaredAnd then we can plot the observed data against the values predicted by our regression to visualize the results. First, remember to tell the notebook that we want our plots to appear in the notebook itself.
%matplotlib inlineimport matplotlib.pyplot as plt
predicted = parameters[0] + parameters[1] * data['temperature'] + parameters[2] * data['rainfall']
plt.plot(predicted, data['mosquitos'], 'ro')
min_mosquitos, max_mosquitos = min(data['mosquitos']), max(data['mosquitos'])
plt.plot([min_mosquitos, max_mosquitos], [min_mosquitos, max_mosquitos], 'k-')[<matplotlib.lines.Line2D at 0x56eb950>]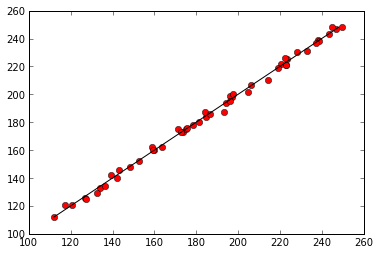
Regression of mosquitoes
OK, great. So putting this all together we now have a piece of code that imports the modules we need, loads the data into memory, fits a regression to the data, and stores the parameters and fit of data.
import pandas as pd
import statsmodels.api as sm
import matplotlib.pyplot as plt
data = pd.read_csv('A2_mosquito_data.csv')
data['temperature'] = (data['temperature'] - 32) * 5 / 9.0
regr_results = sm.OLS.from_formula('mosquitos ~ temperature + rainfall', data).fit()
parameters = regr_results.params
rsquared = regr_results.rsquared
predicted = parameters[0] + parameters[1] * data['temperature'] + parameters[2] * data['rainfall']
plt.plot(predicted, data['mosquitos'], 'ro')
min_mosquitos, max_mosquitos = min(data['mosquitos']), max(data['mosquitos'])
plt.plot([min_mosquitos, max_mosquitos], [min_mosquitos, max_mosquitos], 'k-')
print parameters
print "R^2 = ", rsquaredIntercept 17.545739
temperature 0.871943
rainfall 0.696717
dtype: float64
R^2 = 0.996966873691
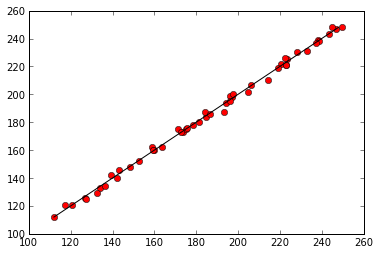
Regression of mosquitoes
Functions
The next thing we need to do is loop over all of the possible data files, but in order to do that we're going to need to grow our code some more. Since our brain can only easily hold 5-7 pieces of information at once, and our code already has more than that many pieces, we need to start breaking our code into manageable sized chunks. This will let us read and understand the code more easily and make it easier to reuse pieces of our code. We'll do this using functions.
Functions in Python take the general form
def function_name(inputs):
do stuff
return outputSo, if we want to write a function that returns the value of a number squared we could use:
def square(x):
x_squared = x ** 2
return x_squared
print "Four squared is", square(4)
print "Five squared is", square(5)Four squared is 16
Five squared is 25
We can also just return the desired value directly.
def square(x):
return x ** 2
print square(3)9
And remember, if we want to use the result of the function later we need to store it somewhere.
two_squared = square(2)
print two_squared4
Challenges
- Write a function that converts temperature from Fahrenheit to Celcius and use it to replace
data['temperature'] = (data['temperature'] - 32) * 5 / 9.0in our program.
- Write a function called
analyze()that takesdataas an input, performs the regression, makes the observed-predicted plot, and returnsparameters.
Walk through someone's result. When discussing talk about different names. E.g., fahr_to_celcius is better than temp_to_celcius since it is explicit both the input and the output. Talk about the fact that even though this doesn't save us any lines of code it's still easier to read.
The call stack
Let's take a closer look at what happens when we call fahr_to_celsius(32.0). To make things clearer, we'll start by putting the initial value 32.0 in a variable and store the final result in one as well:
def fahr_to_celsius(tempF):
tempC = (tempF - 32) * 5 / 9.0
return tempC
original = 32.0
final = fahr_to_celsius(original)Call Stack (Initial State)
When the first three lines of this function are executed the function is created, but nothing happens. The function is like a recipe, it contains the information about how to do something, but it doesn't do so until you explicitly ask it to. We then create the variable original and assign the value 32.0 to it. The values tempF and tempC don't currently exist.
Call Stack Immediately After Function Call
When we call fahr_to_celsius, Python creates another stack frame to hold fahr_to_celsius's variables. Upon creation this stack frame only includes the inputs being passed to the function, so in our case tempF. As the function is executed variables created by the function are stored in the functions stack frame, so tempC is created in the fahr_to_celsius stack frame.
Call Stack At End Of Function Call
When the call to fahr_to_celsius returns a value, Python throws away fahr_to_celsius's stack frame, including all of the variables it contains, and creates a new variable in the original stack frame to hold the temperature in Celsius.
Call Stack After End
This final stack frame is always there; it holds the variables we defined outside the functions in our code. What it doesn't hold is the variables that were in the other stack frames. If we try to get the value of tempF or tempC after our functions have finished running, Python tells us that there's no such thing:
print tempCNameError Traceback (most recent call last)
<ipython-input-14-3054d7679e45> in <module>()
----> 1 print tempC
NameError: name 'tempC' is not definedThe reason for this is encapsulation, and it's one of the key to writing correct, comprehensible programs. A function's job is to turn several operations into one so that we can think about a single function call instead of a dozen or a hundred statements each time we want to do something. That only works if functions don't interfere with each other by potentially changing the same variables; if they do, we have to pay attention to the details once again, which quickly overloads our short-term memory.
Testing Functions
Once we start putting things into functions so that we can re-use them, we need to start testing that those functions are working correctly. The most basic thing we can do is some informal testing to make sure the function is doing what it is supposed to do. To see how to do this, let's write a function to center the values in a dataset prior to conducting statistical analysis. Centering means setting the mean of each variable to be the same value, typically zero.
def center(data):
return data - data.mean()We could test this on our actual data, but since we don't know what the values ought to be, it will be hard to tell if the result was correct. Instead, let's create a made up data frame where we know what the result should look like.
import pandas as pd
test_data = pd.DataFrame([[1, 1], [1, 2]])
print test_data 0 1
0 1 1
1 1 2
Now that we've made some test data we need to figure out what we think the result should be and we need to do this before we run the test. This is important because we are biased to believe that any result we get back is correct, and we want to avoid that bias. This also helps make sure that we are confident in what we want the code to do. So, what should the result of running center(data) be?
OK, let's go ahead and run the function.
print center(test_data) 0 1
0 0 -0.5
1 0 0.5
That looks right, so let's try center on our real data:
data = pd.read_csv('A2_mosquito_data.csv')
print center(data) year temperature rainfall mosquitos
0 -25 1.607843 -7.039216 -5.235294
1 -24 -10.392157 19.960784 8.764706
2 -23 8.607843 23.960784 21.764706
3 -22 -6.392157 -93.039216 -64.235294
4 -21 -2.392157 -60.039216 -45.235294
5 -20 4.607843 -56.039216 -37.235294
6 -19 5.607843 -35.039216 -23.235294
7 -18 -5.392157 -101.039216 -73.235294
8 -17 -10.392157 68.960784 44.764706
9 -16 5.607843 -42.039216 -23.235294
10 -15 2.607843 14.960784 12.764706
11 -14 -2.392157 89.960784 61.764706
12 -13 6.607843 80.960784 62.764706
13 -12 -4.392157 78.960784 53.764706
14 -11 5.607843 23.960784 16.764706
15 -10 9.607843 76.960784 57.764706
16 -9 -4.392157 -17.039216 -10.235294
17 -8 6.607843 49.960784 39.764706
18 -7 7.607843 -79.039216 -52.235294
19 -6 6.607843 10.960784 13.764706
20 -5 0.607843 -1.039216 -1.235294
21 -4 -6.392157 -32.039216 -25.235294
22 -3 4.607843 -5.039216 1.764706
23 -2 -9.392157 -77.039216 -59.235294
24 -1 -0.392157 17.960784 14.764706
25 0 -8.392157 -11.039216 -12.235294
26 1 -4.392157 53.960784 36.764706
27 2 4.607843 -96.039216 -64.235294
28 3 2.607843 39.960784 24.764706
29 4 5.607843 -70.039216 -43.235294
30 5 1.607843 -48.039216 -33.235294
31 6 -3.392157 -35.039216 -25.235294
32 7 -6.392157 72.960784 45.764706
33 8 -10.392157 83.960784 52.764706
34 9 -3.392157 -81.039216 -60.235294
35 10 8.607843 -16.039216 -7.235294
36 11 2.607843 90.960784 62.764706
37 12 -0.392157 74.960784 51.764706
38 13 5.607843 11.960784 9.764706
39 14 -8.392157 -64.039216 -51.235294
40 15 -1.392157 54.960784 35.764706
41 16 4.607843 -18.039216 -10.235294
42 17 5.607843 -2.039216 0.764706
43 18 -8.392157 -12.039216 -12.235294
44 19 -2.392157 -59.039216 -39.235294
45 20 -9.392157 54.960784 33.764706
46 21 7.607843 47.960784 40.764706
47 22 -1.392157 54.960784 35.764706
48 23 -7.392157 -9.039216 -9.235294
49 24 5.607843 7.960784 1.764706
50 25 6.607843 -80.039216 -56.235294
It's hard to tell from the default output whether the result is correct, but there are a few simple tests that will reassure us:
print 'original mean:'
print data.mean()
centered = center(data)
print
print 'mean of centered data:'
centered.mean()original mean:
year 1985.000000
temperature 80.392157
rainfall 207.039216
mosquitos 185.235294
dtype: float64
mean of centered data:
</code></pre><pre class='out'><code>year 0.000000e+00
temperature 1.393221e-15
rainfall 6.687461e-15
mosquitos -1.337492e-14
dtype: float64The mean of the centered data is very close to zero; it's not quite zero because of floating point precision issues. We can even go further and check that the standard deviation hasn't changed (which it shouldn't if we've just centered the data):
print 'std dev before and after:'
print data.std()
print
print centered.std()std dev before and after:
year 14.866069
temperature 6.135400
rainfall 56.560396
mosquitos 39.531551
dtype: float64
year 14.866069
temperature 6.135400
rainfall 56.560396
mosquitos 39.531551
dtype: float64
The standard deviations look the same. It's still possible that our function is wrong, but it seems unlikely enough that we we're probably in good shape for now.
Documentation
OK, the center function seems to be working fine. Does anyone else see anything that's missing before we move on?
Yes, we should write some documentation to remind ourselves later what it's for and how to use it. This function may be fairly straightforward, but in most cases it won't be so easy to remember exactly what a function is doing in a few months. Just imagine looking at our analyze function a few months in the future and trying to remember exactly what it was doing just based on the code.
The usual way to put documentation in code is to add comments like this:
# center(data): return a new DataFrame containing the original data centered around zero.
def center(data, desired):
return data - data.mean()There's a better way to do this in Python. If the first thing in a function is a string that isn't assigned to a variable, that string is attached to the function as its documentation:
def center(data, desired):
"""Return a new DataFrame containing the original data centered around zero."""
return data - data.mean()This is better because we can now ask Python's built-in help system to show us the documentation for the function.
help(center)Help on function center in module __main__:
center(data, desired)
Return a new DataFrame containing the original data centered around zero.
A string like this is called a docstring and there are also automatic documentation generators that use these docstrings to produce documentation for users. We use triple quotes because it allows us to include multiple lines of text and because it is considered good Python style.
def center(data):
"""Return a new array containing the original data centered on zero
Example:
>>> import pandas
>>> data = pandas.DataFrame([[0, 1], [0, 2])
>>> center(data)
0 1
0 0 -0.5
1 0 0.5
"""
return data - data.mean()
help(center)Help on function center in module __main__:
center(data)
Return a new array containing the original data centered on zero
Example:
>>> import pandas
>>> data = pandas.DataFrame([[0, 1], [0, 2])
>>> center(data)
0 1
0 0 -0.5
1 0 0.5
Challenge
- Test your temperature conversion function to make sure it's working (think about some temperatures that you easily know the conversion for).
- Add documentation to both the temperature conversation function and the analysis function.
Looping over files
So now our code looks something like this:
import pandas as pd
import statsmodels.api as sm
import matplotlib.pyplot as plt
def fahr_to_celsius(tempF):
"""Convert fahrenheit to celsius"""
tempC = (tempF - 32) * 5 / 9.0
return tempC
def analyze(data):
"""Perform regression analysis on mosquito data
Takes a dataframe as input that includes columns named 'temperature',
'rainfall', and 'mosquitos'.
Performs a multiple regression to predict the number of mosquitos.
Creates an observed-predicted plot of the result and
returns the parameters of the regression.
"""
regr_results = sm.OLS.from_formula('mosquitos ~ temperature + rainfall', data).fit()
parameters = regr_results.params
predicted = parameters[0] + parameters[1] * data['temperature'] + parameters[2] * data['rainfall']
plt.figure()
plt.plot(predicted, data['mosquitos'], 'ro')
min_mosquitos, max_mosquitos = min(data['mosquitos']), max(data['mosquitos'])
plt.plot([min_mosquitos, max_mosquitos], [min_mosquitos, max_mosquitos], 'k-')
return parameters
data = pd.read_csv('A2_mosquito_data.csv')
data['temperature'] = fahr_to_celsius(data['temperature'])
regr_results = analyze(data)
print parametersIntercept 17.545739
temperature 0.871943
rainfall 0.696717
dtype: float64
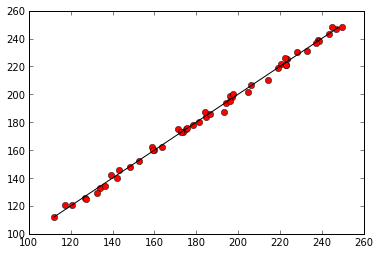
Regression of mosquitoes
Now we want to loop over all of the possible data files, and to do that we need to know their names. If we only had a dozen files we could write them all down, but if we have hundreds of files or the filenames change then that won't really work. Fortunately Python has a built in library to help us find the files we want to work with called glob.
import glob
filenames = glob.glob('*.csv')
print filenames['A1_mosquito_data.csv', 'B2_mosquito_data.csv', 'A3_mosquito_data.csv', 'B1_mosquito_data.csv', 'A2_mosquito_data.csv']
The object returned by glob is a list of strings. A list is a Python data type that holds a group of potentially heterogenous values. That means it can hold pretty much anything, including functions.
mylist = [1, 'a', center]
print mylist[1, 'a', <function center at 0x5c63b18>]
In this case all of the values are strings that contain the names of all of the files that match the expression given to glob, so in this case all of the files with the .csv extension.
Let's restrict the filenames a little more finely, so that we don't accidentally get any data we don't want, and print out the filenames one at a time.
filenames =glob.glob('*data.csv')
for filename in filenames:
print filenameA1_mosquito_data.csv
B2_mosquito_data.csv
A3_mosquito_data.csv
B1_mosquito_data.csv
A2_mosquito_data.csv
Challenge
Modify your code to loop over all of the files in your directory, making an observed-predicted plot for each file and printing the parameters.
